snpboost - Boosting Polygenic Risk Scores
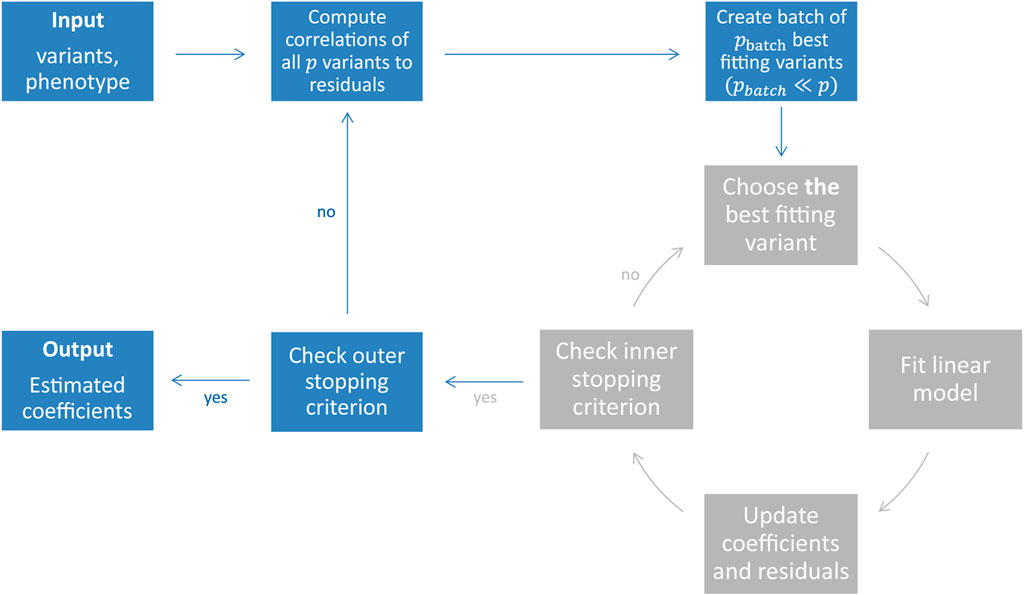
To fit polygenic risk scores (PRS) directly on individual-level genotype data, we developed the adapted statistical boosting framework snpboost which is implemented in R and available open source (https://github.com/hklinkhammer/snpboost). The algorithm is batch-based, i.e. we iteratively work on batches of variants to reduce computational complexity. Statistical boosting fits a multivariable regression, i.e. all genetic information is taken into account simultaneously instead of looking at each SNP separately. While the snpboost framework has been introduced (https://doi.org/10.3389/fgene.2022.1076440), we are currently working on extending the framework to fit different outcomes or phenotypes (including time-to-event outcomes and quantile regression) and to include further effects (e.g. non-linear effects and SNP-SNP interactions). This is joint work with the Institute of Medical Biometry, Informatics and Epidemiology, Bonn University, and the Center for Human Genetics, Philipps-University Marburg.